SARS-CoV-2 Virus
(Redirected from Severe acute respiratory syndrome coronavirus 2)
Jump to navigation
Jump to search
A SARS-CoV-2 Virus is a betacoronavirus with a SARS-CoV-2 genome.
- AKA: Severe Acute Respiratory Syndrome Coronavirus 2.
- Context:
- It can (typically) have a Spike Protein that has a functional polybasic (furin) cleavage at the S1–S2 boundary through the insertion of 12 nucleotides.
- It can be in a SARS-CoV-2 Virus Infected Organism (with a SARS-CoV-2 Viral Load).
- It can (often) cause COVID-19 (COVID-19 cases via COVID-19 infections), such as during the COVID-19 pandemic.
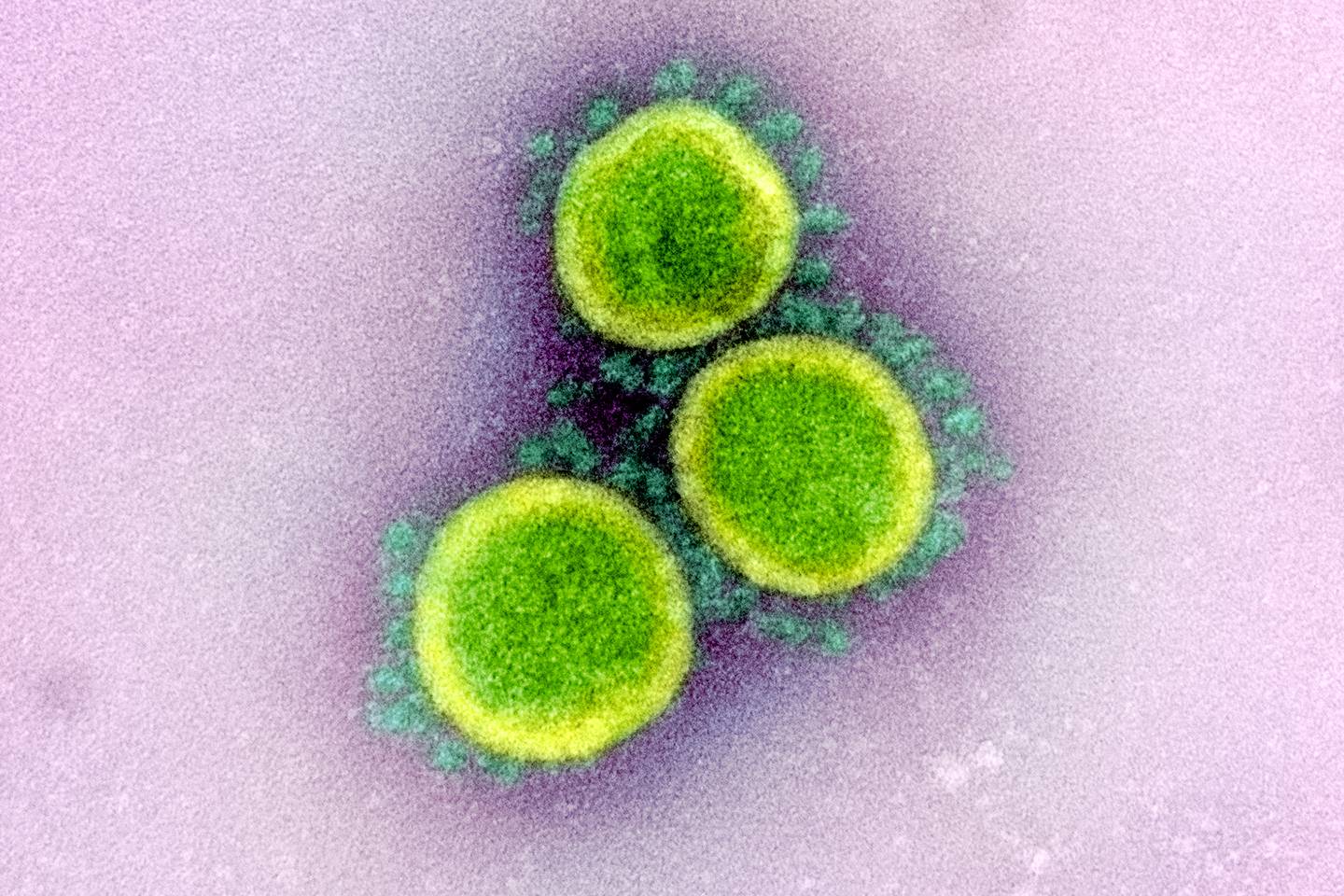
- It can be approximately 50–200 nanometres in diameter.
- …
- Example(s):
- a Human SARS-CoV-2 Virus, such as:
 .
. - A D614G Strain.
- …
- a Human SARS-CoV-2 Virus, such as:
- Counter-Example(s):
- a SARS Virus.
- a Spanish Flu Virus.
- a Flu Virus.
- a Common Cold Virus.
- See: Positive Sense Single-Stranded RNA Virus, Lipid Bilayer.
References
2020
- Muge Cevik. (2020). https://twitter.com/mugecevik/status/1308080067998822401 Sept. 21.
- QUOTE: ... When we look at the viral load dynamics & contact tracing studies, those who are infected are very infectious for a short window, likely 1-2 days before and 5 days following symptom onset. No transmission documented so far after the first week of symptom onset. ...
.
- QUOTE: ... When we look at the viral load dynamics & contact tracing studies, those who are infected are very infectious for a short window, likely 1-2 days before and 5 days following symptom onset. No transmission documented so far after the first week of symptom onset. ...
2020
- (Wikipedia, 2020) ⇒ https://en.wikipedia.org/wiki/Severe_acute_respiratory_syndrome_coronavirus_2 Retrieved:2020-3-1.
- Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), previously known by the provisional name 2019-nCoV, is a positive-sense single-stranded RNA virus. It is contagious in humans and is the cause of coronavirus disease 2019 (COVID-19). SARS-CoV-2 has strong genetic similarity to bat coronaviruses, from which it likely originated, although an intermediate reservoir such as a pangolin is thought to be involved. From a taxonomic perspective SARS-CoV-2 is classified as a strain of the species severe acute respiratory syndrome-related coronavirus. SARS-CoV-2 is the cause of the ongoing 2019–20 coronavirus outbreak, a Public Health Emergency of International Concern that originated in Wuhan, China. Because of this connection, the virus is sometimes referred to informally, among other nicknames, as the "Wuhan coronavirus".
\
2020b
- (Wikipedia, 2020) ⇒ https://en.wikipedia.org/wiki/Severe_acute_respiratory_syndrome_coronavirus_2#Structural_biology Retrieved:2020-3-25.
- Each SARS-CoV-2 virion is approximately 50–200 nanometres in diameter. Like other coronaviruses, SARS-CoV-2 has four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope. The spike protein, which has been imaged at the atomic level using cryogenic electron microscopy, is the protein responsible for allowing the virus to attach to the membrane of a host cell. Protein modeling experiments on the spike protein of the virus soon suggested that SARS-CoV-2 has sufficient affinity to the angiotensin converting enzyme 2 (ACE2) receptors of human cells to use them as a mechanism of cell entry. By 22 January 2020, a group in China working with the full virus genome and a group in the United States using reverse genetics methods independently and experimentally demonstrated that ACE2 could act as the receptor for SARS-CoV-2. Studies have shown that SARS-CoV-2 has a higher affinity to human ACE2 than the original SARS virus strain. SARS-CoV-2 may also use basigin to gain cell entry. Initial spike protein priming by transmembrane protease, serine 2 (TMPRSS2) is essential for entry of SARS-CoV-2. After a SARS-CoV-2 virion attaches to a target cell, the cell's protease TMPRSS2 cuts open the spike protein of the virus, exposing a fusion peptide. The virion then releases RNA into the cell, forcing the cell to produce copies of the virus that are disseminated to infect more cells. SARS-CoV-2 produces at least three virulence factors that promote shedding of new virions from host cells and inhibit immune response.
2020c
- (Andersen et al., 2020) ⇒ Kristian G. Andersen, Andrew Rambaut, W. Ian Lipkin, Edward C. Holmes, and Robert F. Garry. (2020). “The Proximal Origin of SARS-CoV-2.” Nature Medicine. https://doi.org/10.1038/s41591-020-0820-9
- QUOTE: ... Our comparison of alpha- and betacoronaviruses identifies two notable genomic features of SARS-CoV-2: (i) on the basis of structural studies 7,8,9 and biochemical experiments 1,9,10, SARS-CoV-2 appears to be optimized for binding to the human receptor ACE2; and (ii) the spike protein of SARS-CoV-2 has a functional polybasic (furin) cleavage site at the S1–S2 boundary through the insertion of 12 nucleotides8, which additionally led to the predicted acquisition of three O-linked glycans around the site. ...