CogStack Ecosystem
A CogStack Ecosystem is a information processing system designed for working with EHR databases.
- Context:
- Website: https://cogstack.org
- Source code repository: https://github.com/CogStack
- It has been implemented as part of the SemEHR Producing Subsystem.
- It has been developed as an open source project with the source code available at: https://github.com/CogStack/ .
- It is composed of the following microservices:
- CogStack Samples Database (
samples-db) - PostgreSQL database loaded with a sample dataset; - CogStack Pipeline (
cogstack-pipeline) - CogStack data processing engine; - CogStack Job Repository (
cogstack-job-repo)– PostgreSQL database for storing information about CogStack jobs, - CogStack-ElasticSearch (
elasticsearch-1) – ElasticSearch search engine (single node) for storing and querying the processed EHR data, - Kibana UI (
kibana) - Kibana data visualization tool for querying the data from ElasticSearch.
- CogStack Samples Database (
- It can be deployed by with many running services we refer as an ecosystem or a platform
- Example(s):
- CogStack-NiFi,
- Processing a simple, structured dataset from a single DB source.
- Processing a combined structured and free-text dataset from a single DB source.
- Processing a combined dataset from multiple DB sources, multiple jobs.
- Processing a combined dataset with embedded documents from a single DB source.
- 2-step processing of a combined dataset with embedded documents from a single DB source.
- Basic security use-case: Example 2 extended reverse proxy enabling a secure access.
- Logging: Example 6 extended with logging mechanisms.
- Simple NLP use-case: drug annotation using GATE.
- Defining multi-component pipelines.
- …
- Counter-Example(s):
- See: Docker, Data Processing Pipeline, SemEHR System, Semantic Web Search System, Semantic Web, Ontology Search System, Natural Language Processing, Annotation Task, Electronic Health Record.
References
2019a
- (CogStack Doc., 2019) ⇒ https://cogstack.atlassian.net/wiki/spaces/COGDOC/overview Retrieved: 2019-04-27.
- QUOTE: CogStack is a lightweight distributed, fault tolerant database processing architecture and ecosystem, intended to make NLP processing and preprocessing easier in resource constrained environments. It comprises of multiple components, where CogStack Pipeline, the one covered in this documentation, has been designed to provide a configurable data processing pipelines for working with EHR data. For the moment it mainly uses databases and files as the primary source of EHR data with the possibility of adding custom data connectors in the near future. It makes use of the Java Spring Batch framework in order to provide a fully configurable data processing pipeline with the goal of generating an annotated JSON files that can be readily indexed into ElasticSearch, stored as files or pushed back to a database.
The CogStack ecosystem has been developed as an open source project with the code available on GitHub: https://github.com/CogStack/ .
- QUOTE: CogStack is a lightweight distributed, fault tolerant database processing architecture and ecosystem, intended to make NLP processing and preprocessing easier in resource constrained environments. It comprises of multiple components, where CogStack Pipeline, the one covered in this documentation, has been designed to provide a configurable data processing pipelines for working with EHR data. For the moment it mainly uses databases and files as the primary source of EHR data with the possibility of adding custom data connectors in the near future. It makes use of the Java Spring Batch framework in order to provide a fully configurable data processing pipeline with the goal of generating an annotated JSON files that can be readily indexed into ElasticSearch, stored as files or pushed back to a database.
2019b
- (Cogstack Doc., 2019) ⇒ https://cogstack.atlassian.net/wiki/spaces/COGDOC/pages/38043684/Quickstart Retrieved: 2019-04-27.
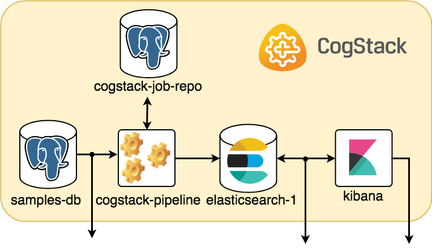
- QUOTE: CogStack ecosystem: The picture below sketches a general idea on how the microservices are running and communicating within a sample CogStack ecosystem used in this tutorial.
Assuming that everything is working fine, we should be able to connect to the running microservices. Retrieved: 2019-04-27.
- QUOTE: CogStack ecosystem: The picture below sketches a general idea on how the microservices are running and communicating within a sample CogStack ecosystem used in this tutorial.
2019c
- (Roguski, 2019) ⇒ Lukasz Roguski (Jan, 2019). "CogStack platform"
- QUOTE: In this part are covered the available services that can be running in an example CogStack deployment. To such deployment with many running services we refer as an ecosystem or a platform. Below is presented a high-level perspective of CogStack platform with the possibilities it enables through many components and services. (...) In practice, many of the functionalities that CogStack platform enables are implemented as separate, but interconnected services working inside the ecosystem. (...) In most scenarios CogStack platform will consist of core services tailored to specific use-cases. Additional application and services can be run on top of it, such as SemEHR, Patient Timeline, Live Alerting (through ElasticSearch plugins) or any other custom developed applications. For an ease-of-use, when deploying a sample CogStack platform, we always emphasise to use Docker Compose (see: Running CogStack).
Below is presented is one of the most simple and common scenarios when ingesting and processing the EHR data from a proprietary data source.
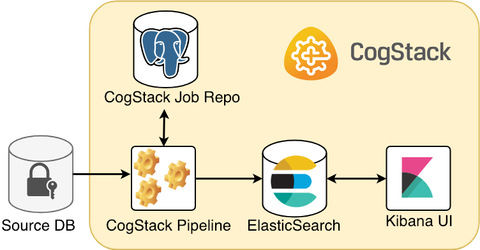
A CogStack platform presented here consists of such core services:
- CogStack Pipeline service for ingesting and processing the EHR data from the source database,
- CogStack Job Repository (PostgreSQL database) serving for job status control,
- ElasticSearch sink where the processed EHR records are stored,
- (optional) Kibana user interface to easily perform exploratory data analysis over the processed records.
- QUOTE: In this part are covered the available services that can be running in an example CogStack deployment. To such deployment with many running services we refer as an ecosystem or a platform. Below is presented a high-level perspective of CogStack platform with the possibilities it enables through many components and services. (...) In practice, many of the functionalities that CogStack platform enables are implemented as separate, but interconnected services working inside the ecosystem. (...) In most scenarios CogStack platform will consist of core services tailored to specific use-cases. Additional application and services can be run on top of it, such as SemEHR, Patient Timeline, Live Alerting (through ElasticSearch plugins) or any other custom developed applications. For an ease-of-use, when deploying a sample CogStack platform, we always emphasise to use Docker Compose (see: Running CogStack).
2018a
- (Jackson et al., 2018) ⇒ Richard Jackson, Ismail Kartoglu, Clive Stringer, Genevieve Gorrell, Angus Roberts, Xingyi Song, Honghan Wu, Asha Agrawal, Kenneth Lui, Tudor Groza, Damian Lewsley, Doug Northwood, Amos Folarin, Robert Stewart, and Richard Dobson. (2018). “CogStack - Experiences of Deploying Integrated Information Retrieval and Extraction Services in a Large National Health Service Foundation Trust Hospital.” In: BMC Medical Informatics and Decision Making Journal, 18(47). doi:10.1186/s12911-018-0623-9
- QUOTE: CogStack is a set of open source and open core services, co-ordinated by a batch processing framework that builds on the concepts of the Cognition platform by offering additional interfaces for NHS systems and NLP technologies. Out-of-the-box open source components were selected from a variety of successful open source and freely licensed projects. The services can be deployed using the Docker containerisation technology, to maximise ease of deployment.
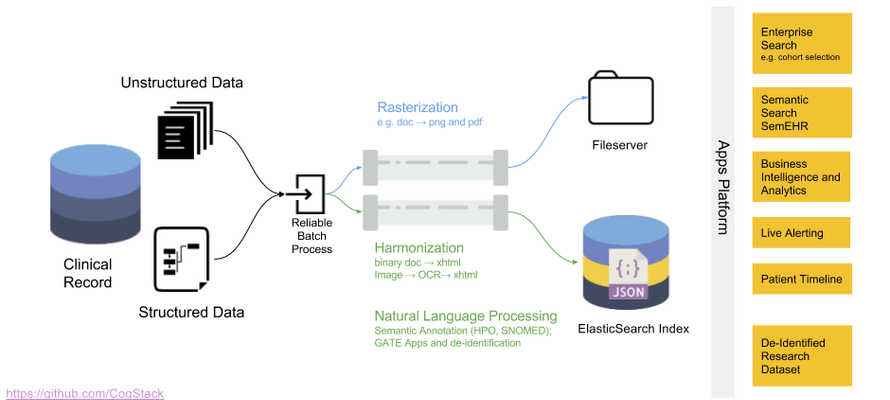
The overall goal of the architecture is to undertake a series of configurable transformations of clinical data housed in relational databases and to load the transformed data into an Elasticsearch information retrieval engine (otherwise known as a search engine - described below), whereupon the 3R principles can be more readily addressed than via direct communication with the untransformed source databases alone. Each transformation is highly configurable, in accordance with the desired use case of the end product. For example, it is not necessary (or even desirable) to de-identify data for business intelligence use cases, and thus this can be disabled. Similarly, not all use cases will require computationally expensive entity extraction NLP processes. The rationale for the choice of components is described below, while the flow of data and transformations in the CogStack architecture is described in Fig. 1.
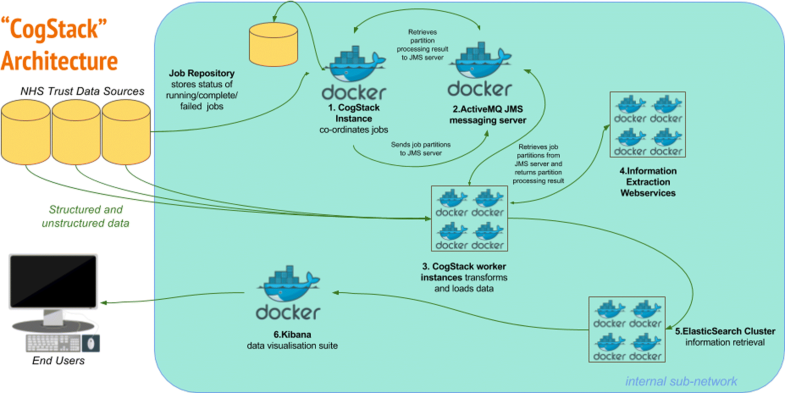
Fig. 1: CogStack Architecture and Dataflow. All components can be deployed via the Docker containerisation software. 1. New job execution Master instance of CogStack identifies new data in Trust Data Sources at intermittent intervals. 2. Partitioning The job is partitioned into a user definable number of work units. 3a. Derive the freetext content Extract plain and/or formatted text from common proprietary document binary formats (performing OCR where necessary), using the Tika Library to enable the downstream processing of high value unstructured data elements. 3b. Supplement the text content with meta-data Filter and de-normalise a subset of the structured clinical data to provide a patient orientated, transparent representation of high value metadata concepts. For example, this might include calculated fields to represent patient age at document date, first part of postcode and ethnicity and lab results. 3c. De-identification Transform the resulting text documents into de-identified text documents, by masking personal health identifiers via the use of the Cognition de-identification algorithms. This is necessary to address governance concerns associated with the secondary use of patient data. Identifiers in structured data can be excluded via SQL query, according to business requirements. 4. Information Extraction Apply generic clinical IE pipelines to derive additional structured data from free text and supplement the quantity of available structured data at the point of query. 5. Indexing Build a JSON object from the resulting structured and unstructured data, which can then be readily be indexed into an Elasticsearch cluster. 6. Visualisation The Kibana suite provides a range of attractive options for viewing, aggregating and dash-boarding the loaded data.
- QUOTE: CogStack is a set of open source and open core services, co-ordinated by a batch processing framework that builds on the concepts of the Cognition platform by offering additional interfaces for NHS systems and NLP technologies. Out-of-the-box open source components were selected from a variety of successful open source and freely licensed projects. The services can be deployed using the Docker containerisation technology, to maximise ease of deployment.
2018b
- (Wu et al., 2018) ⇒ Honghan Wu, Giulia Toti, Katherine I Morley, Zina M Ibrahim, Amos Folarin, Richard Jackson, Ismail Kartoglu, Asha Agrawal, Clive Stringer, Darren Gale, Genevieve Gorrell, Angus Roberts, Matthew Broadbent, Robert Stewart, and Richard JB Dobson. (2018). “SemEHR: A General-purpose Semantic Search System to Surface Semantic Data from Clinical Notes for Tailored Care, Trial Recruitment, and Clinical Research.” In: Journal of the American Medical Informatics Association, 25(5).
2017
- (Wu et al., 2017) ⇒ Honghan Wu, Giulia Toti, Katherine I Morley, Zina Ibrahim, Amos Folarin, Ismail Kartoglu, Richard Jackson, Asha Agrawal, Clive Stringer, Darren Gale, Genevieve M Gorrell, Angus Roberts, Matthew Broadbent, Robert Stewart, and Richard J B Dobson. (2017). “SemEHR: Surfacing Semantic Data from Clinical Notes in Electronic Health Records for Tailored Care, Trial Recruitment, and Clinical Research.” In: The Lancet, 390. doi:10.1016/S0140-6736(17)33032-5